RNase H2 Enzyme
Virtually eliminate primer-dimers in PCR
Increase the precision of target sequence amplification by limiting negative effects from primer-dimers. For use in customer-developed rhPCR assay applications, such as rare variant identification, splice variant discovery, or microbial identification studies.
Ordering
- Reduce the formation of primer-dimers or misprimed PCR products
- Reliably multiplex a large number of DNA targets
In addition to your choice of Taq DNA polymerase and other general PCR reagents, rhPCR technology requires the following components:
- rhPCR Primers—use instead of conventional PCR primers
- RNase H2 Enzyme—needed for activation of rhPCR primers and PCR product extension
Product details
rhPCR calls for specially designed primers (rhPCR Primers) and a thermostable RNase H2 Enzyme. We offer RNase H2 Enzyme at two concentrations, 20 U/µL and 2 U/µL.* RNase H2 is functional in most PCR buffers and can be added directly to your PCR master mix. The enzyme must be diluted before use with the RNase Enzyme Dilution Buffer supplied with the enzyme. Detergent is required for ideal Pyrococcus abyssi RNase H2 activity. The IDT dilution and storage buffer for RNase H2 contains 0.1% Triton™ X-100.
RNase H2 Enzyme is an endogenous endoribonuclease that binds to RNA-DNA duplexes and cleaves the RNA strand, leaving a 5' phosphate and a 3' hydroxyl group. RNase H2 will cleave at a single ribonucleotide residue embedded within a heteroduplex. RNase H2 will not cleave single-stranded RNA.
*One unit is defined as the amount of enzyme necessary to nick 1 nmol of synthetic, single-rC-containing duplex substrate per minute at 70°C in 10 mM Tris, 50 mM NaCl, 0.01% Triton X-100, 10 µg/ml BSA, 4 mM MgCl2. Depending on the application, enzyme usage range is 2−200 mU per reaction.
Product data
How rhPCR works
Sometimes for PCR, it is necessary to position primers at suboptimal locations in the target. This can result in the formation of primer-dimers and/or undesired amplification of homologous sequences.
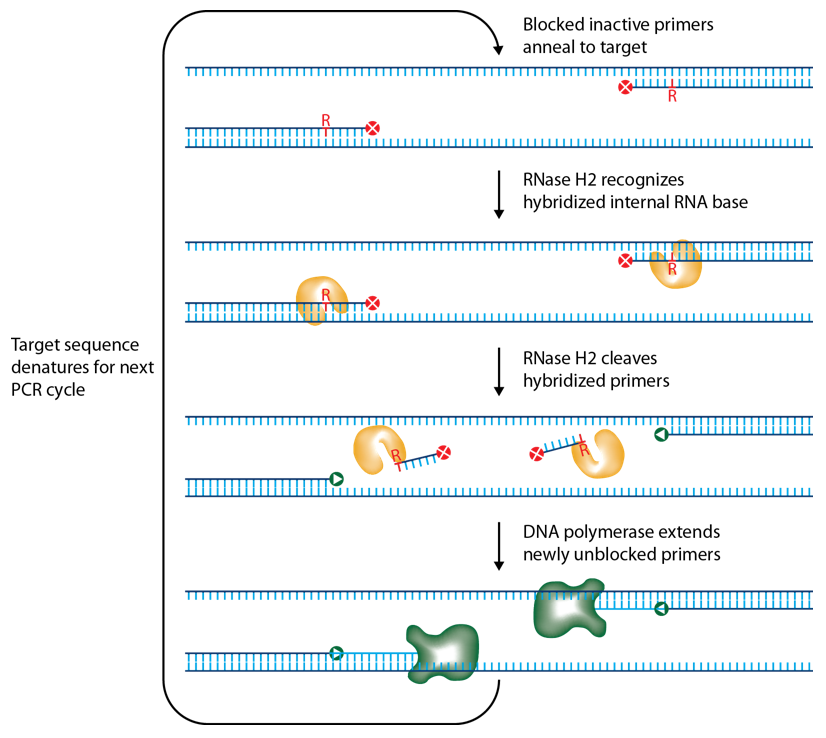
IDT scientists have developed RNase H2-dependent PCR (rhPCR), a method for increasing PCR precision and reducing the formation of primer-dimers by using RNase H2 from Pyrococcus abyssi (P. abyssi) and DNA primers that contain a single ribonucleotide residue and a 3' blocking moiety. The blocked primers are activated when cleaved by the RNase H2 Enzyme. Cleavage occurs on the 5' side of the RNA base after primer hybridization to the target DNA. Because the primers can only be cleaved after they hybridize to the perfectly matched target sequence, primer-dimers are reduced. The requirement for high target complementarity reduces amplification of closely related sequences (Figure 1).
P. abyssi is a hyperthermophile, so the P. abyssi RNase H2 Enzyme has optimal activity in range of 70–75°C and is functional in rhPCR between 50−75°C. P. abyssi RNase H2 has very low activity at room temperature (~1000X less activity) [1]. Therefore, use of this enzyme for primer activation confers a "hot start" characteristic to the PCR. P. abyssi RNase H2 is functional in most PCR buffers and can be added directly to your PCR master mix.
rhPCR largely reduces primer-dimer artifacts
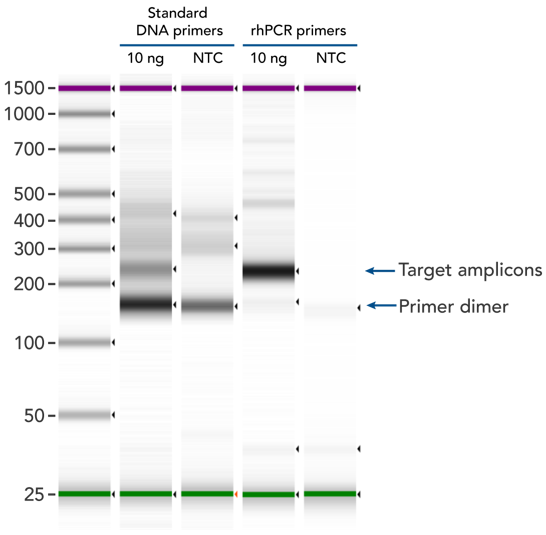
rhPCR uses activation of RNA-DNA hybrid primers by the RNase H2 Enzyme from P. abyssi. The primers are only activated upon precise annealing to the correct target, and this RNase H2 activation mechanism reduces primer-dimers and other amplification artifacts that can affect experimental results. Figure 2 shows how effectively RNase H2 activation reduces primer-dimer artifacts, even when 96 primer pairs are multiplexed in a single PCR.
Figure 2. rhPCR primers largely reduce primer-dimers and nonspecific amplification artifacts in multiplex applications. In multiplex PCR amplification of 96 targets from human genomic DNA (NA12878, Coriell Institute), 2 sets of multiplex primers for the 96 assays (192 individual primers) were synthesized either as standard PCR primers or as RNase H2-activated, rhPCR primer pairs. The results show tight control of primer-dimer artifacts, even in highly multiplexed assays. Total reaction volumes were 50 µL, with 10 ng of genomic DNA template or no template as controls. RNase H2 was present in all reactions but has no functional role in PCRs with standard primers.
Thermostability
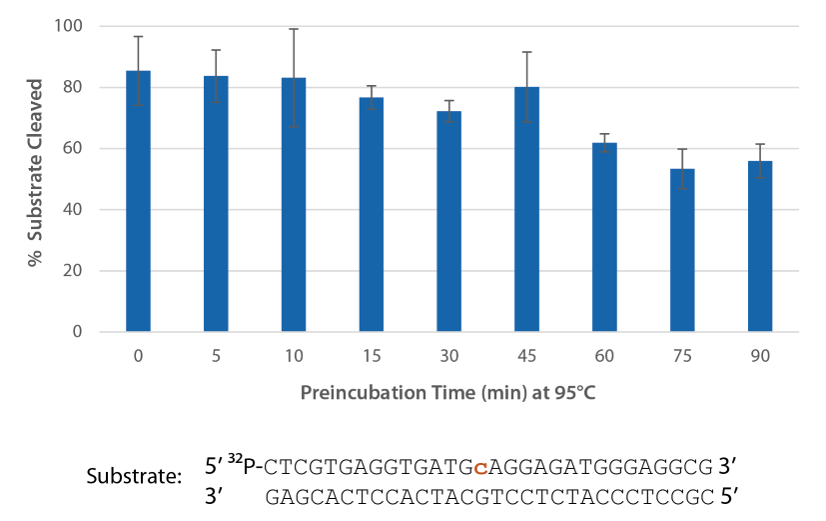
P. abyssi RNase H2 can be incubated at 95°C for >45 minutes with little loss of activity (Figure 3) and, therefore, can survive PCR conditions.
Figure 3. Thermostability of P. abyssi RNase H2 allows this enzyme to survive PCR conditions. P. abyssi RNase H2 was preincubated at 95°C for 0−90 min in buffer (10 mM Tris HCl, pH 8.0; 50 mM NaCl; 0.01% Triton X-100; 10 µg/mL BSA; and 4 mM MgCl2). Radiolabeled substrate (2 pmol) was added to heat-treated RNase H2 (100 µU), and enzyme activity was assessed by measuring substrate cleavage after incubation at 70°C for 20 min. Error bars represent standard deviation (n = 3).
Compatibility with PCR temperatures
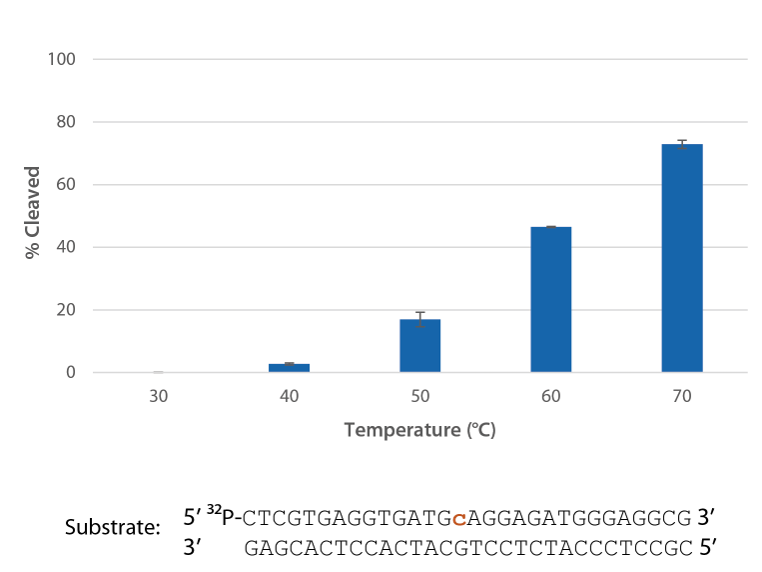
P. abyssi RNase H2 is active at temperatures that are compatible with PCR conditions and inactive at room temperature (Figure 4).
- P. abyssi RNase H2 can be used to create "hot start" PCR conditions without the need for a modified "hot start" polymerase, because RNase H2 is essentially inactive at room temperature.
- P. abyssi RNase H2 activity rises slow after 70°C, and greatly decreases at 80°C (data not shown).
- Compared to other RNases, P. abyssi RNase H2 is relatively safe to have in laboratories that work with RNA, because this RNase H2 requires elevated temperatures, Mg2+, detergents, and most importantly, duplexed RNA/DNA substrate for activity.
Figure 4. P. abyssi RNase H2 is active at temperatures compatible with PCR and inactive at room temperature (20−25°C). Enzyme activity of P. abyssi RNase H2 (250 µU) was assessed by measuring cleavage of a radiolabeled substrate (2 pmol) after incubation at varying temperatures for 20 min in buffer (10 mM Tris HCl, pH 8.0; 50 mM NaCl; 0.01% Triton X-100; 10 µg/mL BSA; and 4 mM MgCl2). Error bars represent standard deviation (n = 3).
Compatibility with various Mg2+ concentrations used in PCR
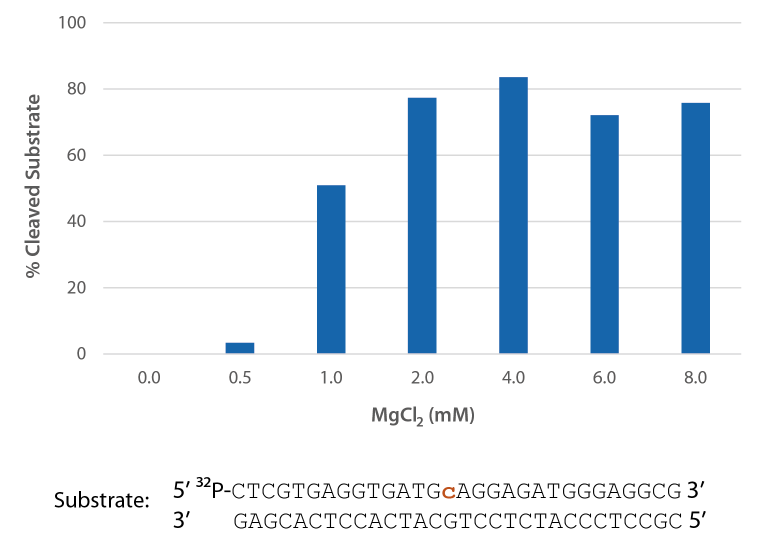
P. abyssi RNase H2 is active across a broad range of Mg2+ concentrations (Figure 5) that are compatible with PCR conditions (typically 3 mM).
Figure 5. P. abyssi is active at Mg2+ concentrations compatible with PCR. Enzyme activity of P. abyssi RNase H2 (250 µU) was assessed by measuring cleavage of a radiolabeled substrate (2 pmol) after incubation at 70°C for 20 min in buffer (10 mM Tris HCl, pH 8.0; 50 mM NaCl; 0.01% Triton X-100; and 10 µg/mL BSA), containing 0–8 mM MgCl2 (n = 2 per condition).
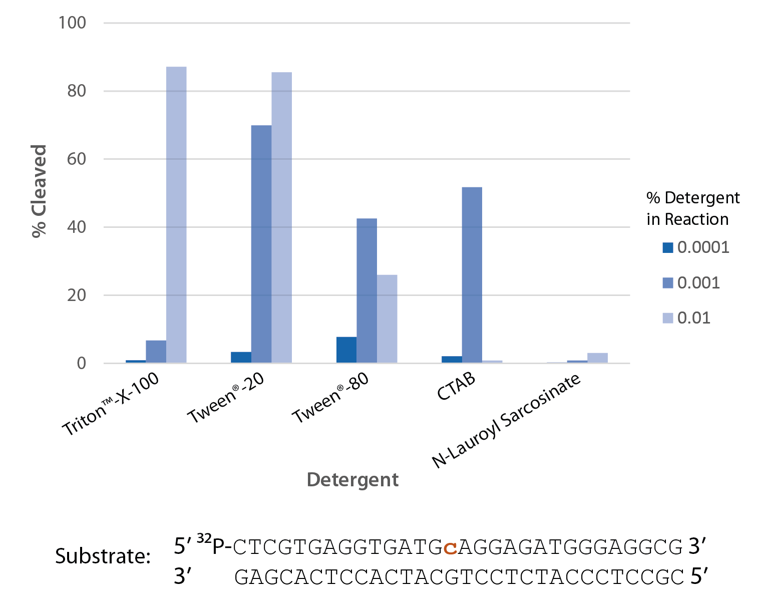
Detergent is required for optimal enzyme activity
The highest enzyme activity in reactions with the detergents tested is observed with Triton X-100 and Tween® 20 (Figure 6).
Figure 6. P. abyssi RNase H2 activity requires detergent. Enzyme activity of P. abyssi RNase H2 (100 µU) was assessed by measuring cleavage of a radiolabeled substrate (2 pmol) after incubation at 70°C for 30 min in buffer (10 mM Tris HCl, pH 8.0; 50 mM NaCl; 10 µg/mL BSA; and 4 mM MgCl2), containing varying amounts of different detergents (n = 1 per condition).
Resources
Related products
References
- Dobosy JR, Rose SD, Beltz KR, et al. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol. 2011;11:80.